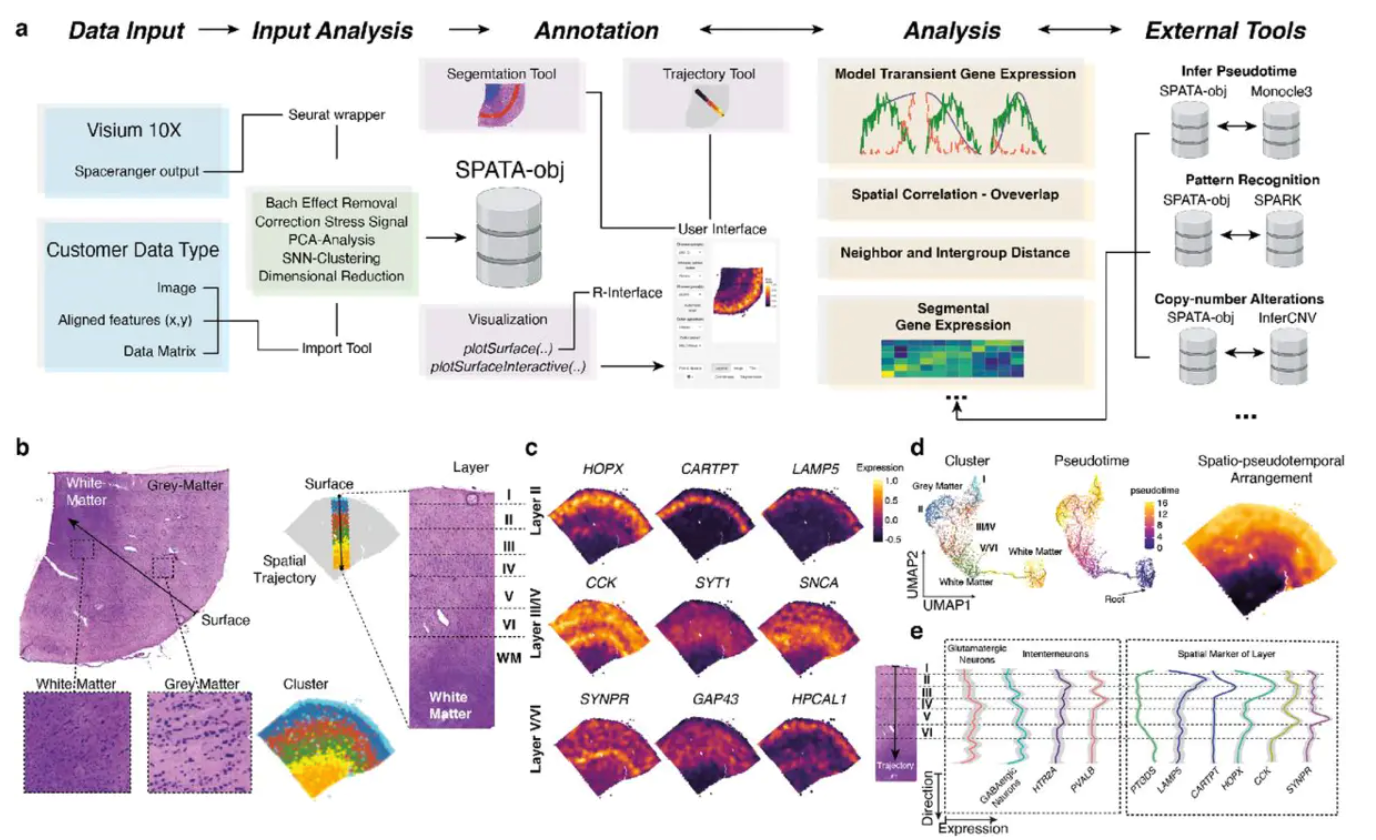
SPATA - SPAtial Transcriptomic Analysis : 空间转录组分析-提供了一个R函数和交互应用的工具,使空间转录数据的分析更容易。SPATA提供易于使用的界面、易于可视化数据的简单函数,也为有经验的R用户提供的更复杂的和自由的函数。我们知道,高通量转录组数据分析落脚点往往是一个基因集,有了这个基因集我们可以富集出通路,查看调控关系等。以往的分析往往是用一种方法先得到一个基因集再用下一个工具来研究基因集,为什么不直接把基因集的背景知识放到空间转录组的数据分析框架之中呢?SPATA就是在做这样的一个探索。
SPATA 哲学
第一篇空转的论文发表以来,已经发布了一些专门研究空间转录组数据分析和可视化的r包。尽管所有这些包都有其自身的优点和缺点,但我们发现始终难以将我们自己的想法集成到它们预定义的工作流中,这最终促使我们创建了SPATA。虽然SPATA引入了自己新的空间分析方法——比如空间轨迹建模——同时也提供了已有的可视化工具——比如表面绘图(surface plotting)——可以将SPATA看作一个更通用的框架,可以方便地集成用户的想法。通过提供这样一个框架,如提供易于使用、交互式的应用程序、对函数和方法的全面了解,不管研究者的编程技能如何,都可以快速上手空转分析。直接的对象转换函数允许在任何时间点从SPATA切换到不同的数据分析平台,如Seurat或Monocle(反之亦然),从而易于利用和集成所有包的分析优势。
在所有的函数和应用程序中,SPATA都实现了数据整理(tidy-data)的概念,它的主要理念来自于tidyverse。这意味着SPATA中的所有数据框架都面向整齐的数据结构,其中每一行代表一个观察,每一列代表该观察的一个变量(信息)。一致的输出和一致的术语通过强大的tidyverse和tidymodeling函数促进了个人想法的应用。对于空间转录组数据的分析,我们称之为spata-data.frame的每一个观察都代表一个条形码点,而归属变量(列)代表与这些点相关的额外信息。
条形码和坐标能够唯一地识别每个spot,这一事实乍一看似乎是显而易见的,几乎没有人对此感兴趣。然而,从数据科学的角度来看,它拥有巨大的力量,因为这些关键变量允许您轻松地集成无限个维度来分析。条形码序列允许集成数千个基因和基因集表达水平,而坐标则允许集成所有类型的数据以及坐标,如成像数据、空间细胞数据等。尽管空间转录组实验伴随着空间维度的基因和基因集表达数据,仍然在某种程度上类似于任何其他测序技术的数据,一种技术的方法适用于另一种技术。优秀的如Seurat利用了这一点,并提供了各种工具来集成空间和单细胞数据。除了它自己新的可视化和分析工具之外,SPATA还尝试方便地集成与坐标一起出现的各种数据。此外,它允许您将spata对象与Seurat和Monocle3之间自由转换,进一步拓展数据探索空间。
在这里,我们不再完全演示SPATA的所有功能,而是只演示演示者觉得能够体现SPATA特征的功能。具体地,SPATA可以做什么:
- Data Visualization
- Genes, Gene-sets & Features
- Spatial Segmentation & Clustering
- Differential Expression Analysis
- Spatial Trajectory Analysis
- Extract, join & add data
- Integrate your ideas
- Integrate different platforms
基因集可视化
library(SPATA)
library(magrittr)
library(ggplot2)
library(patchwork)
# create a spata-object from scratch
spata_obj <-
initiateSpataObject_10X(input_paths = c("F:\\EE\\learnscanpy\\data\\V1_Breast_Cancer_Block_A_Section_1_spatial\\"), sample_names = c("BCBS"))
#以下是运行提示
No gene-set data.frame path specified.
Step 1/6 : Reading in .h5 file.
Loading from directory: 'F:\EE\learnscanpy\data\V1_Breast_Cancer_Block_A_Section_1_spatial\/outs'
Step 2/6: Extracting count matrices.
Step 3/6: Extracting images and coordinates-information.
Step 4/6: Performing Seurat-analysis steps.
Skip running SCTransform as it's argument input is neither TRUE nor a list.
Step 5/6: Initiating spata-object.
Using SPATA's default gene set data.frame.
Skipping step 6/6 as 'output_path' was set to NULL.
Done.
可见在创建spata-object的时候,已经内置地把Seurat的标准流程跑完了,快速地进入了以空间特征为主要内容的挖掘方向。
几乎没有人能在一天之内完成单细胞数据分析,所以我们先设置保存环境以方便以后直接读入。
# if you want to save a spata object from your session use
saveSpataObject(object = spata_obj,
output_path = "F:\\EE\\learnscanpy\\data\\V1_Breast_Cancer_Block_A_Section_1_spatial\\", # e.g. a folder named 'data'
file_name = "example2")
grep("example2",list.files("F:\\EE\\learnscanpy\\data\\V1_Breast_Cancer_Block_A_Section_1_spatial\\"),value=T)
[1] "spata-obj-example2.RDS"
每遇到一个数据对象,我们多需要看看它是什么样子的。
str(spata_obj,max.level = 2)
Formal class 'spata' [package "SPATA"] with 11 slots
..@ coordinates :'data.frame': 3813 obs. of 4 variables:
..@ data :Formal class 'data_counts' [package "SPATA"] with 2 slots
..@ dim_red :Formal class 'dim_red' [package "SPATA"] with 2 slots
..@ fdata :'data.frame': 3813 obs. of 9 variables:
..@ image :List of 1
..@ samples : chr "BCBS"
..@ scvelo : list()
..@ used_genesets:'data.frame': 936495 obs. of 2 variables:
..@ trajectories :List of 1 # 质控分群注释终于和轨迹放到一起了
..@ version :List of 4
..@ additional :List of 1
head(spata_obj@fdata)
barcodes sample nCount_RNA nFeature_RNA percent.mt percent.RB RNA_snn_res.0.8 seurat_clusters segment
1 AAACAAGTATCTCCCA-1_BCBS BCBS 8399 3795 4.427678 4.695721 0 0
2 AAACACCAATAACTGC-1_BCBS BCBS 37258 8226 4.372967 6.048173 1 1
3 AAACAGAGCGACTCCT-1_BCBS BCBS 2726 1828 2.815594 4.146040 0 0
4 AAACAGGGTCTATATT-1_BCBS BCBS 15963 5167 2.614412 6.700379 0 0
5 AAACAGTGTTCCTGGG-1_BCBS BCBS 36212 8015 4.052591 6.116451 1 1
6 AAACATTTCCCGGATT-1_BCBS BCBS 21092 6615 2.344805 4.321316 2 2
也许是出于希望给空间赋予更多意义,SPATA内置了一个通路基因集。富集分析在我们传统的基因集分析种事很常见的,这里把它内置这里,以便我们直接分析基因集(score)。
getGeneSetOverview(spata_obj)
Class Available Gene Sets
1 BC 289
2 BP.GO 7269
3 CC.GO 972
4 HM 50
5 MF.GO 1581
6 RCTM 1493
length(unique(spata_obj@used_genesets$ont)) # getGeneSets(spata_obj)
[1] 11654
head(unique(spata_obj@used_genesets$ont))
[1] "BC_RELA_PATHWAY" "BC_NO1_PATHWAY" "BC_CSK_PATHWAY" "BC_SRCTMRPTP_PATHWAY" "BC_AMI_PATHWAY"
[6] "BC_GRANULOCYTES_PATHWAY"
查看分群结果与基因的可视化
# plot a cluster feature
p1 <-
plotSurface(object = spata_obj,
of_sample = "BCBS",
color_to = "seurat_clusters",
pt_clrp = "npg",
pt_size = 1.5) + labs(color = "Clusters")
# plot gene-set expression
p2 <-
plotSurface(object = spata_obj,
of_sample = "BCBS",
color_to = "CARTPT",
pt_size = 1.5,
pt_clrsp = "magma")
# plot gene-set expression (spatially smoothed)
p3 <-
plotSurface(object = spata_obj,
color_to = "CARTPT",
pt_size = 1.6,
pt_clrsp = "magma",
smooth = TRUE,
smooth_span = 0.01)
# combine with patchwork
p1 +
theme(legend.position = "none") +
p2 +
theme(legend.position = "none")+
p3 +
theme(legend.position = "top")
plotSurface函数用于在空间上绘制信息,我们看到最右侧的图对基因表达做了空间平滑,要比中间的图好看的多。空转的可视化确实是个难题啊。绘制基因集score的score是哪来的呢?
?plotSurfaceComparison
Character value. The method according to which gene sets will be handled specified as a character of length one. This can be either 'mean' or one of 'gsva', 'ssgsea', 'zscore', or 'plage'. The latter four will be given to gsva::GSVA().
可以看到其实就是我们常用的基因集打分方法。下面来可视化一下吧:
?plotSurfaceComparison
p5 <- plotSurfaceComparison(object = spata_obj,
variables = "RCTM_FBXW7_MUTANTS_AND_NOTCH1_IN_CANCER",
smooth = TRUE,
smooth_span = 0.02,
pt_size = 1,
pt_clrsp = "inferno")
# genes_of_interest <- c("CARTPT", "OLIG1", "GFAP", "SYNPR", "HOPX", "CCK")
# e.g. violinplot
?plotDistribution
p6<- plotDistribution(spata_obj,
variables = grep("CANCER",(unique(spata_obj@used_genesets$ont)),value = T)[1:5],
plot_type = "violin",) +
theme(axis.text.x = element_text(angle = 90, hjust = 1,size = 0.3)) # = adjust layout to compensate for long gene set names
# e.g. density
p7<- plotDistribution(spata_obj,
variables = grep("CANCER",(unique(spata_obj@used_genesets$ont)),value = T)[1:5],
plot_type = "density")
p6+p5+ p7

?plotDistributionAcross
genes_of_interest <- c("CARTPT", "OLIG1", "GFAP", "HOPX")
plotDistributionAcross(object = spata_obj,
variables = genes_of_interest,
across = "seurat_clusters",
plot_type = "violin",
clrp = "npg",
ncol = 1) +scale_fill_manual(values=colors())+
theme(legend.position = "none")
基于spot的聚类结果:
p7<- plotSurface(object = spata_obj,
color_to = "seurat_clusters",
pt_clrp = "npg") +
theme(legend.position = "top") +
labs(color = "Seurat Clusters")
p8<- plotUMAP(spata_obj, color_to = "seurat_clusters", pt_alpha = 0.8) +
theme(legend.position = "none") +
labs(color = "Seurat Clusters",
caption = "UMAP")
p9<- plotTSNE(spata_obj, color_to = "seurat_clusters", pt_alpha = 0.8) +
theme(legend.position = "top") +
labs(color = "Seurat Clusters",
caption = "TSNE")
p7 + p8 + p9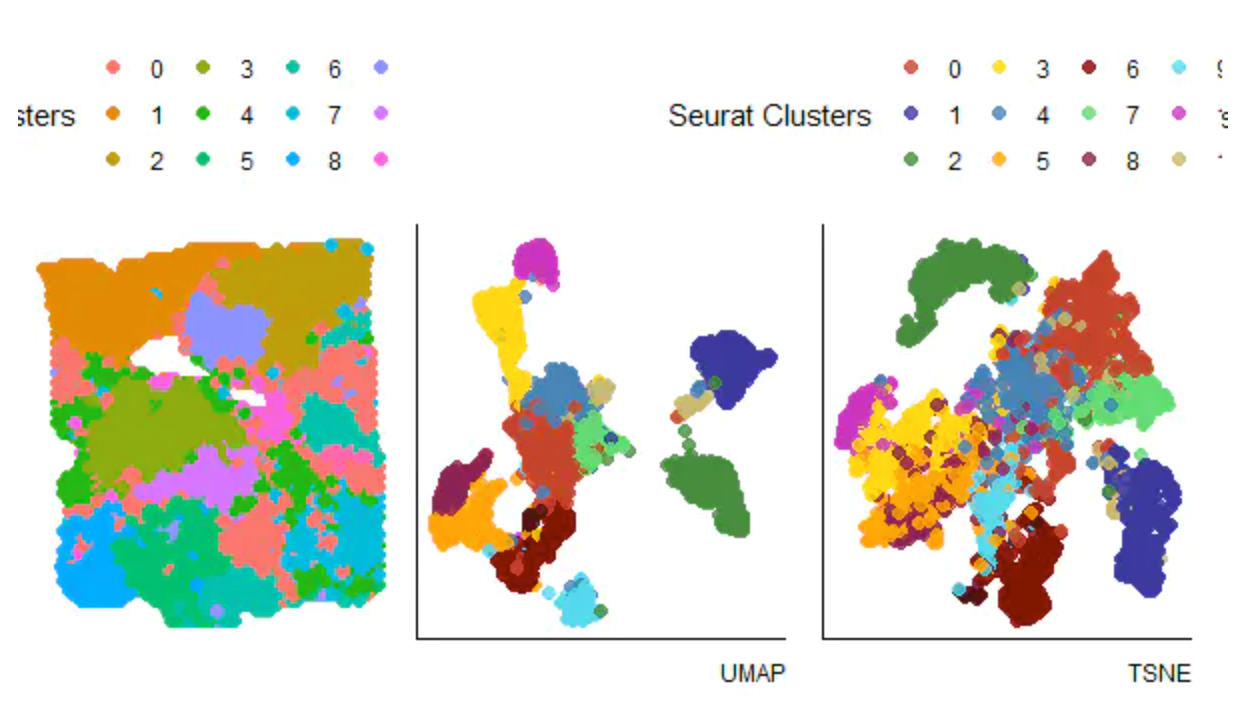
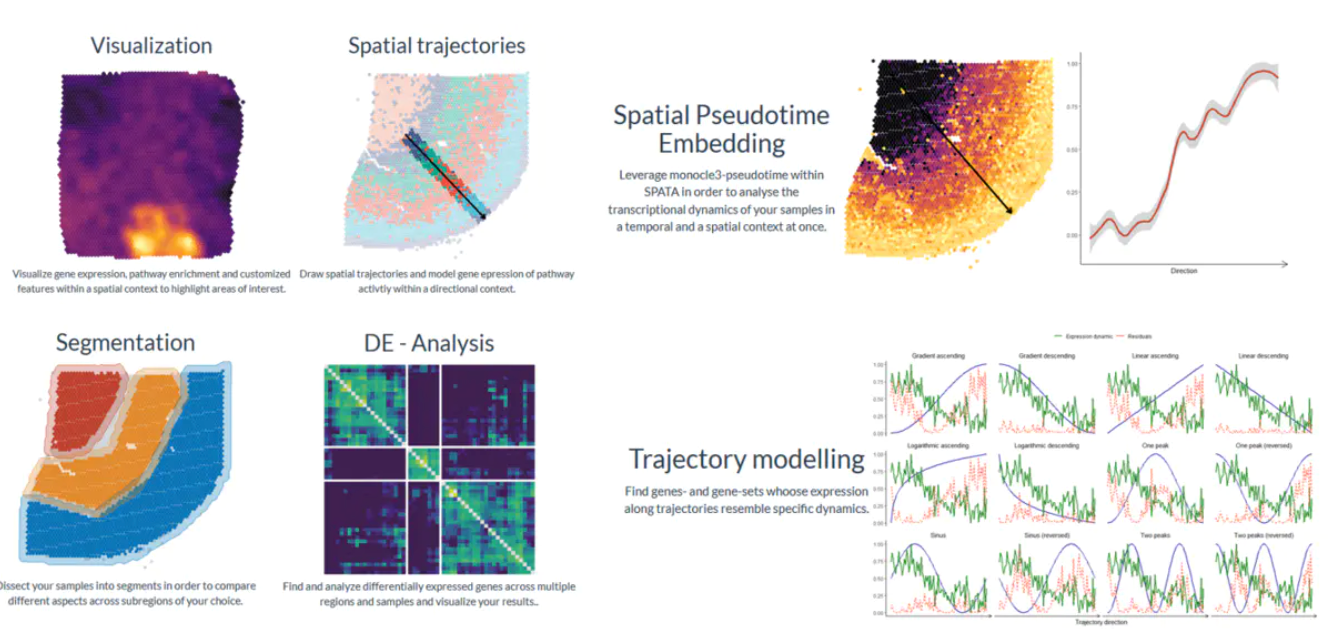
空间轨迹推断
通过空间轨迹分析,SPATA引入了一种新的方法来发现、分析和可视化空间背景下不同表达的基因和基因集。而经典的差异基因表达分析整体实验组之间的差异,它忽略了只有在保持空间维度时才能看到的表达水平的变化。空间轨迹可以回答包含空间分量的问题。例如:
- 我们越靠近感兴趣的区域,表达水平的变化会有多大?
- 哪些基因沿着这些路径遵循相同的模式?
# load package
library(SPATA)
library(magrittr)
library(ggplot2)
library(patchwork)
给定的spata对象中的样本的空间轨迹可以使用函数createTrajectories()交互绘制,如下面的示例所示。[createTrajectories()](https://themilolab.github.io/SPATA/reference/createTrajectories.html)
打开一个shiny应用程序。该应用程序允许一方面像plotSurfaceInteractive()那样调查与空间基因表达有关的样本,另一方面通过四个简单的步骤绘制感兴趣的领域的轨迹。
spata_obj <- createTrajectories(object = spata_obj)
head(getFeatureData(object = spata_obj) )
barcodes sample nCount_RNA nFeature_RNA percent.mt percent.RB RNA_snn_res.0.8 seurat_clusters segment
1 AAACAAGTATCTCCCA-1_BCBS BCBS 8399 3795 4.427678 4.695721 0 0
2 AAACACCAATAACTGC-1_BCBS BCBS 37258 8226 4.372967 6.048173 1 1
3 AAACAGAGCGACTCCT-1_BCBS BCBS 2726 1828 2.815594 4.146040 0 0
4 AAACAGGGTCTATATT-1_BCBS BCBS 15963 5167 2.614412 6.700379 0 0
5 AAACAGTGTTCCTGGG-1_BCBS BCBS 36212 8015 4.052591 6.116451 1 1
6 AAACATTTCCCGGATT-1_BCBS BCBS 21092 6615 2.344805 4.321316 2 2
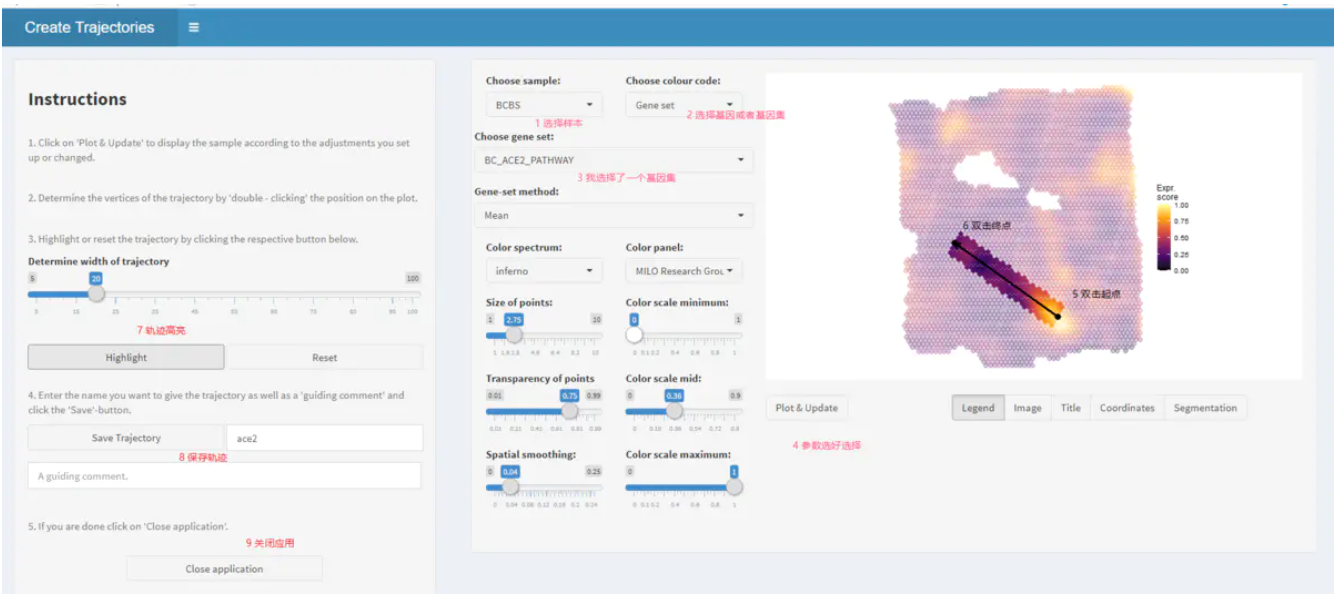
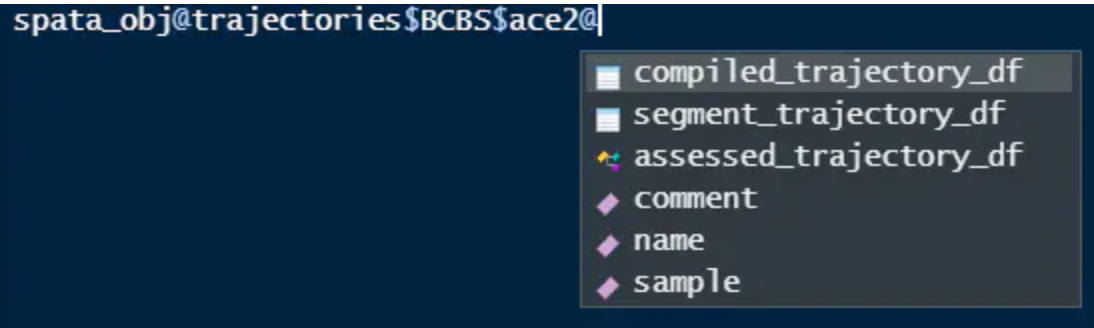
接下来,我们可以看到在空间分群中这个条自定义的轨迹是如何指向的。
plotTrajectory(object = spata_obj,
trajectory_name = "ace2",
color_to = "seurat_clusters",
pt_size = 1.7,
pt_clrp = "npg",
display_image = FALSE) +
theme(legend.position = "top") 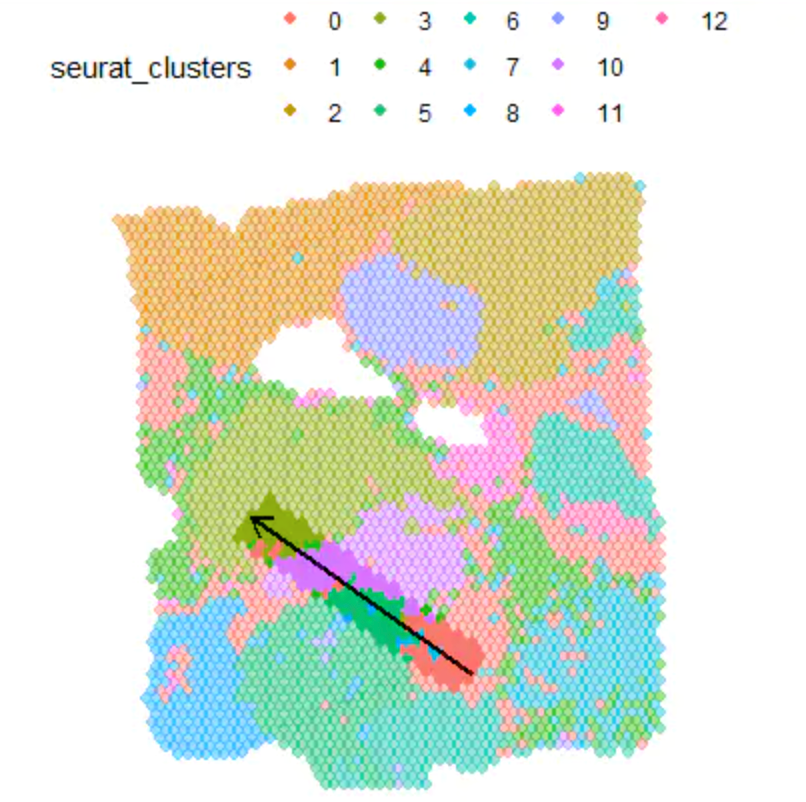
离散变量可以这样绘制:
plotTrajectoryFeaturesDiscrete(object = spata_obj,
trajectory_name = "ace2",
discrete_feature = "seurat_clusters",
clrp = "npg",
display_trajectory_parts = FALSE)
指定感兴趣的基因或基因集。
# gene-set names
genes_of_interest <- c("CALM1", "VIM")
# plot lineplot
plotTrajectoryGenes(object = spata_obj,
trajectory_name = "ace2",
genes = genes_of_interest,
smooth_span = 0.2,
plotTrajectoryGeneSets(
object = spata_obj,
trajectory_name = "ace2",
gene_sets = "HM_HYPOXIA",
display_trajectory_parts = FALSE) + # results in missing vertical lines
ggplot2::theme(legend.position = "top")
使用assessTrajectoryTrends()来查找沿着兴趣轨迹遵循某种趋势的变量。它将所有指定变量的轨迹趋势拟合成各种模式,并通过计算残差曲线下的面积来评估其拟合情况。
all_genes <- getGenes(spata_obj)
all_gene_sets <- getGeneSets(spata_obj)
# obtain an assessed trajectory data.frame for all genes
atdf_gs <- assessTrajectoryTrends(object = spata_obj,
trajectory_name = "ace2",
variables = all_genes)
# obtain an assessed trajectory data.frame for all gene-sets
atdf_gene_sets <- assessTrajectoryTrends(object = spata_obj,
trajectory_name = "ace2",
variables = all_gene_sets[1:5]) # 为了节约内存只做了5个
# output example
atdf_gs
# A tibble: 249,172 x 3
# Groups: variables [17,798]
variables pattern auc
<chr> <chr> <dbl>
1 MYL9 Logarithmic descending 0.516
2 AEBP1 Logarithmic descending 0.523
3 COL1A1 Logarithmic descending 0.527
4 BGN Logarithmic descending 0.543
5 LGALS1 Logarithmic descending 0.681
6 EMILIN1 Logarithmic descending 0.692
7 APOD Logarithmic descending 0.753
8 CAVIN1 Logarithmic descending 0.765
9 ALB Logarithmic descending 0.781
10 SFRP2 Logarithmic descending 0.803
# ... with 249,162 more rows
atdf_gene_sets
# A tibble: 70 x 3
# Groups: variables [5]
variables pattern auc
<chr> <chr> <dbl>
1 BC_AMI_PATHWAY Logarithmic descending 0.980
2 BC_AMI_PATHWAY Linear descending 3.18
3 BC_NO1_PATHWAY Logarithmic descending 3.90
4 BC_NO1_PATHWAY Linear descending 4.03
5 BC_AMI_PATHWAY Gradient descending 4.22
6 BC_SRCTMRPTP_PATHWAY Linear ascending 4.43
7 BC_CSK_PATHWAY Logarithmic descending 4.47
8 BC_RELA_PATHWAY Linear descending 4.48
9 BC_NO1_PATHWAY Gradient descending 5.18
10 BC_CSK_PATHWAY One peak (reversed) 5.23
# ... with 60 more rows
plottrajecyfit()可视化一个特定变量的拟合结果。
# compare the trend of a variable to different models
plotTrajectoryFit(object = spata_obj,
trajectory_name = "ace2",
variable = "MYL9",
display_residuals = TRUE) +
ggplot2::theme(legend.position = "top")
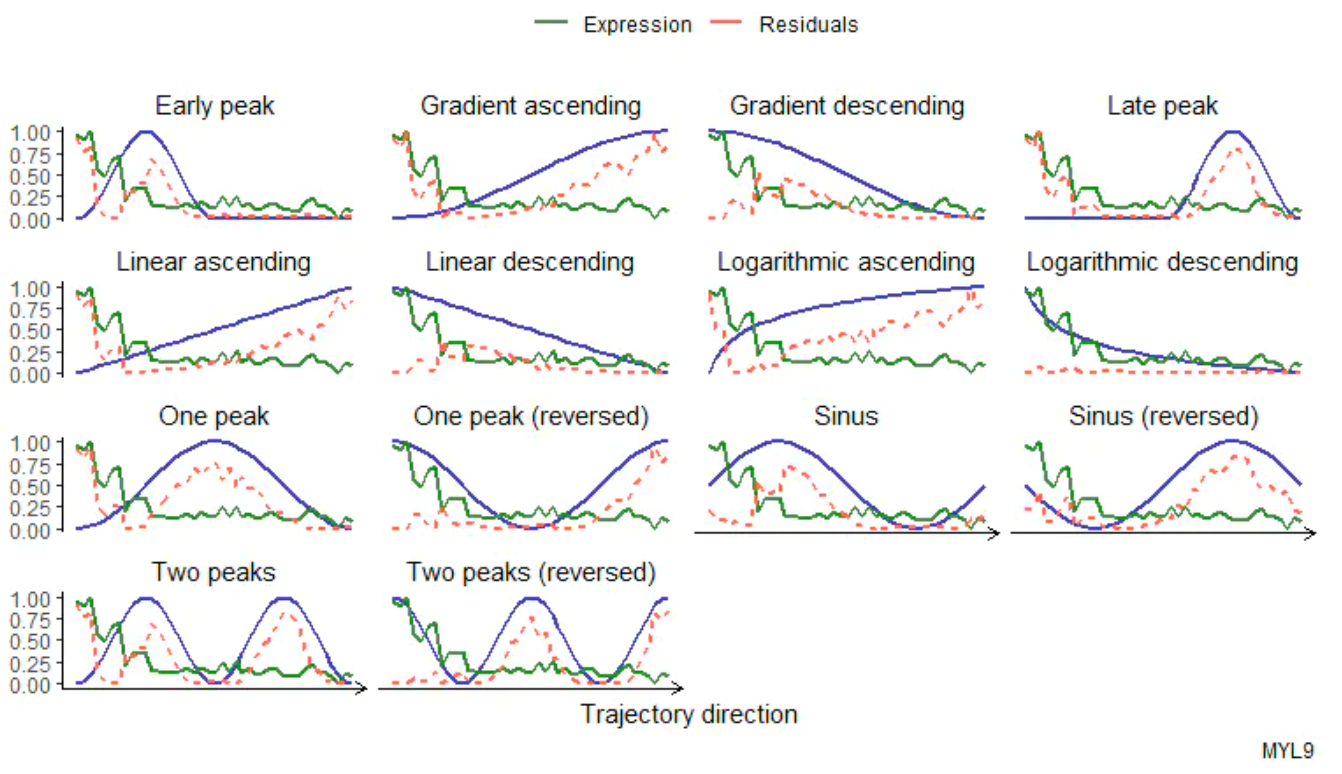
要得到轨迹评估的总结概览,使用examineTrajectoryAssessment()函数显示测量曲线下所有区域的分布。使用filterTrend()或dplyr::filter()和dplyr::pull()的组合评估轨迹数据。
descending_gene_sets <-
filterTrends(atdf = atdf_gene_sets,
limit = 2,
trends = c("Logarithmic descending"),
variables_only = FALSE) # return a data.frame
descending_gene_sets
# A tibble: 1 x 3
# Groups: pattern [1]
variables pattern auc
<chr> <chr> <dbl>
1 BC_AMI_PATHWAY Logarithmic descending 0.980
寻找沿轨迹变化的基因
peaking_genes <- filterTrends(atdf = atdf_gs,
limit = 4,
trends = c("Early peak", "Late peak", "One peak"),
variables_only = TRUE) # return a vector of variables
head(peaking_genes)
[1] "PTMA" "SERF2" "S100A14" "KRT8" "CDH1" "SLC39A6"
在直线图中显示太多变量的表达趋势并不是一个很好的可视化策略。为了处理多个变量,使用plottrajecyheatmap()函数。虽然轨迹目录仍然水平显示,但基因集的动态表达值现在是通过一行颜色的逐渐变化来显示,而不是线图中直线的斜率。当需要显示的变量数量增加时,使用热图来显示沿轨迹的动态表达式变化的优势就变得明显起来。
descending_gene_sets <- dplyr::pull(descending_gene_sets, variables)
plotTrajectoryHeatmap(object = spata_obj,
trajectory_name = "ace2",
variables = descending_gene_sets,
arrange_rows = "maxima",
show_row_names = FALSE, # gene set names are long...
split_columns = TRUE,
smooth_span = 0.5)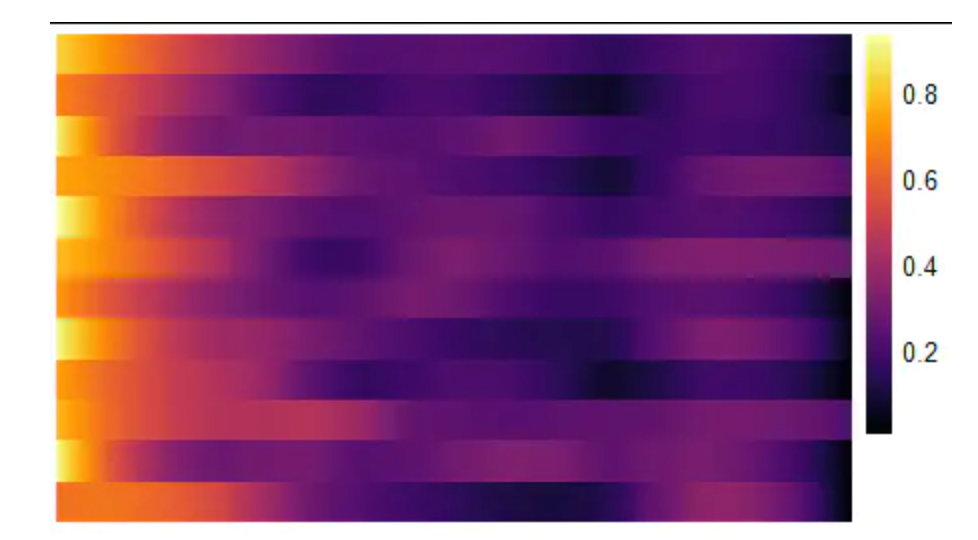
plotTrajectoryHeatmap(object = spata_obj,
trajectory_name = "ace2",
variables = peaking_genes,
arrange_rows = "maxima",
show_row_names = FALSE, # gene set names are long...
split_columns = TRUE, # splits the heatmap
smooth_span = 0.5)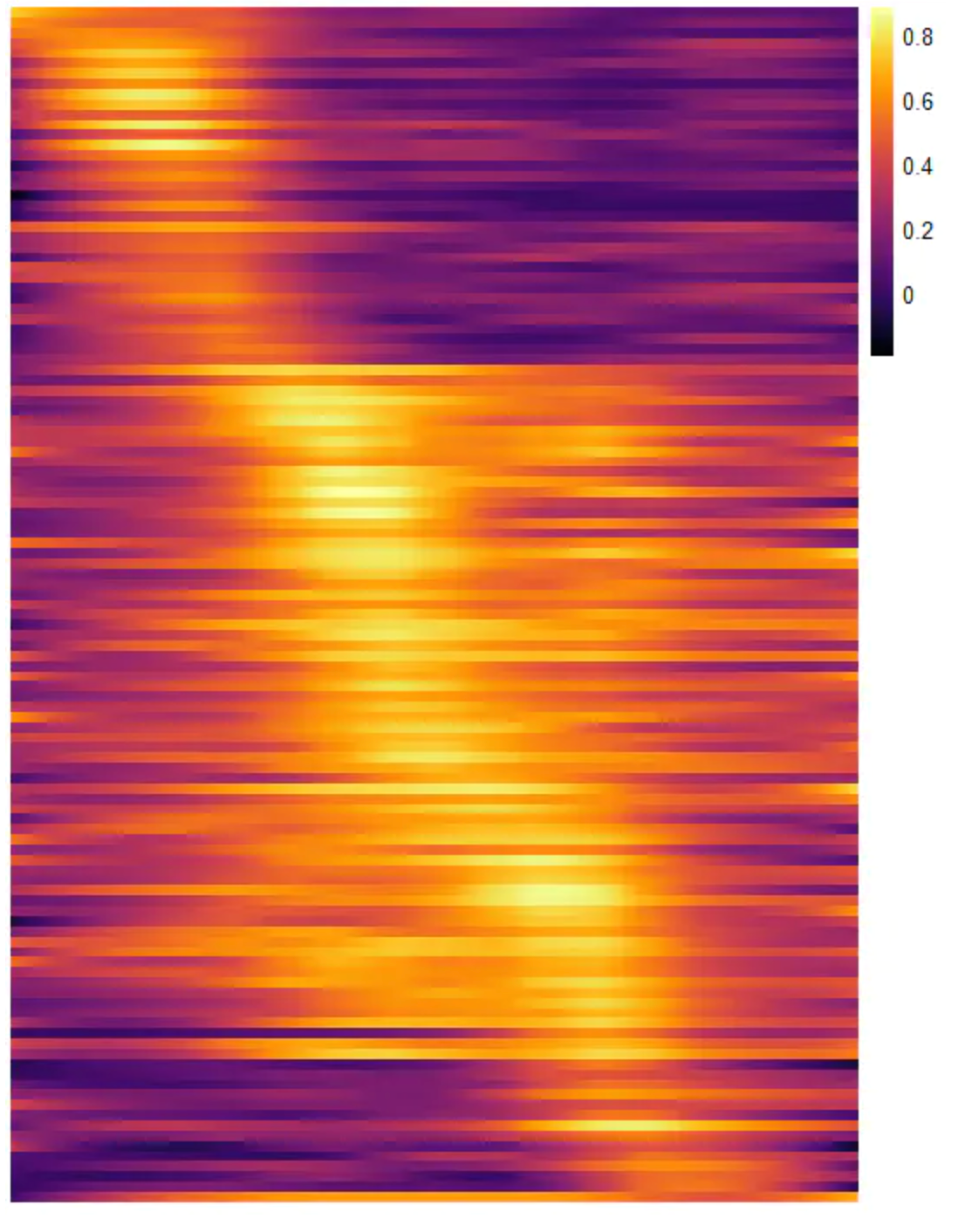
https://themilolab.com/
Inferring spatially transient gene expression pattern from spatial transcriptomic studies
本网站所有内容来源注明为“梅斯医学”或“MedSci原创”的文字、图片和音视频资料,版权均属于梅斯医学所有。非经授权,任何媒体、网站或个人不得转载,授权转载时须注明来源为“梅斯医学”。其它来源的文章系转载文章,或“梅斯号”自媒体发布的文章,仅系出于传递更多信息之目的,本站仅负责审核内容合规,其内容不代表本站立场,本站不负责内容的准确性和版权。如果存在侵权、或不希望被转载的媒体或个人可与我们联系,我们将立即进行删除处理。
在此留言









#SpA#
38
#转录#
37
#转录组分析#
36
#转录组#
34
#ATA#
34